We use mathematical analysis and computer simulations to understand the spread and particularly the control of plant and tree diseases. Our theoretical work attempts to isolate the ways in which factors including host growth, host topography, pathogen dispersal, asymptomatic infection and biological control affect the pattern of spread. However, we have also been involved in developing large-scale, spatially-explicit, stochastic, simulation models that can be fitted to data on the real-world spread of pathogens of current regulatory concern. Examples include sudden oak death, Chalara ash dieback, Dutch elm disease, citrus canker and huanglongbing. This type of model can be used to accurately predict the risk of disease in a given region and/or to quantify the likely effect of any proposed control strategy, together with its inherent risk of failure. Projects include:
Landscape scale epidemiological modelling
Plant pathogens spread at the scale of countries or even continents, and models must reflect this. Working at these scales requires efficient computation, and careful use of (often patchy) spread data. We work on dynamic simulation models of a number of pathogens, including diseases of both trees and crops.
Fungicide resistance and effective use of host plant resistance genes
Anthropomorphic control of crop disease exerts strong selective pressures on plant pathogens. In turn this leads to a risk of breakdown of control, as pathogens become resistant to chemicals or acquire virulence, allowing infection of resistant hosts. We are using mathematical modelling to develop: i) fungicide application strategies to delay the build-up of fungicide resistance; and ii) deployment strategies for host resistance genes which reduce the impact of virulent pathogens.
Optimising controls
Optimising control strategies is difficult, because which strategies will be effective depends on a complex interplay between the epidemiology of the plant-pathogen interaction, the current state of the epidemic and the logistics of detection and control. We have developed an interactive interface, available at http://www.webidemics.com/, designed to illustrate some of these issues to an audience of stakeholders and policy makers. Current work focuses on identifying and optimising control for a range of systems. As well as chemical and genetic controls, this includes cultural and biological disease controls. There is also work on optimal control theory, showing how simplified models can be coupled to complex simulation models to develop control strategies of practical relevance that are informed by solid mathematical theory.
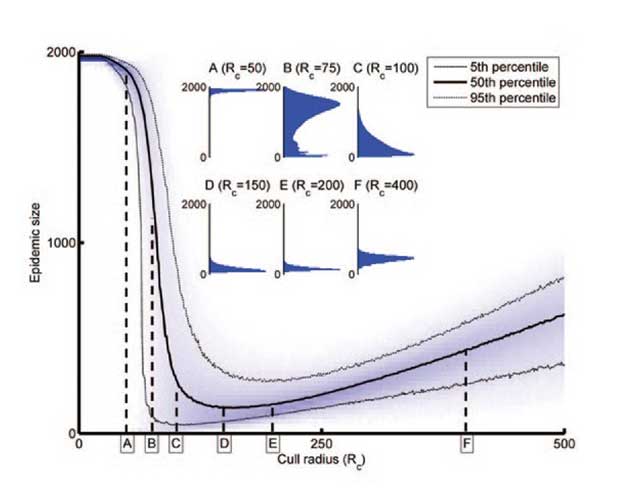
Control fast or control smart?
It is intuitively clear that any attempt to control invasive disease is most likely to succeed when any epidemic remains small, particularly in cases where control targets localised eradication. However, at the start of an epidemic little will be known about the dynamics of pathogen spread and few data are available, and so the impact of control strategies is difficult to assess and optimise. There is therefore an unfortunate but unavoidable trade-off between acting quickly but with limited knowledge, and acting slowly, which means more knowledge is available, but that there is also a larger problem to tackle. Following initial work in the area, we continue to investigate the idea using models of a range of systems.
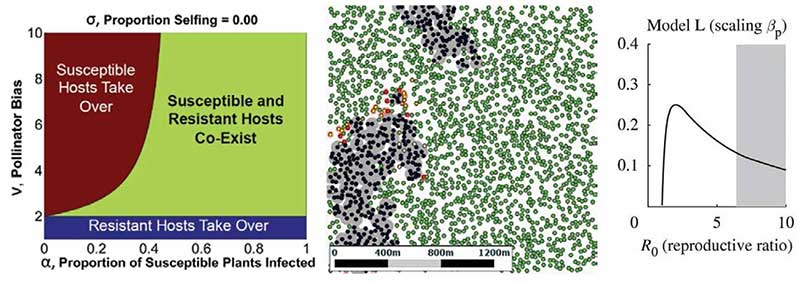
Left: Evolutionary modelling can track genetics of virus resistance. Middle: User friendly spatial models reveal why control strategies can sometimes fail. Right: Increasing the amount of control can lead to a paradoxical increase in the prevalence of disease.