As part of our celebrations to mark 300 years since the appointment of the first Professor of Botany, some of our current academics have written short research stories to help give you an insight into current areas of interest and future research challenges.
If you are interested in finding out more, including how you might be able to support our academics in their future research endeavours, please get in touch with them directly.
Exploring the genetic and epigenetic structure of plant genomes
Ian Henderson, Head of Genetic and Epigenetic Inheritance in Plants Group
Why are plant genomes so large?
Plant genomes are notoriously complex and repetitive, and many are significantly larger than the human genome. For example, Paris japonica has a genome size of ~149 Gigabase (Gb; where 1 Gigabase = 1 billion nucleotides), compared to our own more modestly sized ~3 Gb genome. This apparent disconnect between genome size and perceived organismal complexity is termed the C-value paradox, where the C-value refers to the amount of DNA per genome.
We are interested in the mechanisms and forces that shape plant genomes and allow them to grow into such large and complex architectures. This includes the role of so-called repetitive ‘selfish DNA’. These enigmatic sequences include transposable elements that were identified by Barbara McClintock as jumping genes. Despite being regarded as ‘parasitic’ sequences, increasing amounts of data indicate that sequence repeats can play key functional roles in chromosome biology, evolution, and plant phenotypes. As many of our most important crop species have highly complex and repetitive genomes, it is important to understand the regulation of repeated sequences, as we adapt crops to the changing climate.
Illuminating the dark matter of plant genomes
Highly repetitive regions of the genome have been challenging to study, as previous sequencing methods could only resolve short fragments around one hundred nucleotides in length. Piecing these short fragments together into a complete genome map represents a kind of very challenging jigsaw puzzle, which was essentially impossible to confidently resolve in many cases. However, in recent years, we have experienced a revolution in genome science, with new long-read technologies emerging. These new, much longer sequencing reads - up to hundreds of thousands of nucleotides in length - mean we can now accurately assemble complex repeat regions for the first time, finally illuminating the dark matter of the genome. This means that complex sequences, for example, the large populations of transposable elements that reside in plant genomes, can be studied in new ways.
Why do repeated sequences evolve so fast?
A further mystery surrounding repeated sequences is that they often show very rapid rates of evolutionary change compared to gene-rich regions. For example, in recent years, we have explored the structure and organisation of the centromere regions using long-read sequencing. Centromeres are vital regions of plant and animal chromosomes that connect to the spindle microtubules during cell division and pull them to opposite cell poles. Despite this conserved function, the centromeres are changing very rapidly during evolution. We are currently exploring why this happens, from the perspective of the recombination pathways that act in the centromeres, and also whether competition between selfish centromeres can explain their rapid divergence. As specific transposable element families also reside in the centromeres, we are interested to understand their regulation and what their place is within the centromere paradox. It is very timely to ask these questions due to the new availability of long read sequencing tools that allow us to ask more precise questions and obtain more robust answers about how repeated sequences evolve.
What are the open questions we would like to answer?
Using long-reads we have recently gained sight of previously obscure regions of plant genomes. Understanding how these newly uncovered regions function and evolve is of major interest, including the role of epigenetic information, as repetitive sequences are frequently densely DNA methylated and thereby silenced. Most of our work to date has explored genome regulation in the model plant Arabidopsis thaliana, which has a relatively small genome size (~130 megabases) and is highly experimentally tractable. However, in the future, we would like to expand our studies to other plant systems to explore conserved and contrasting modes of genome evolution. For example, we have an emerging project examining somatic genome evolution in ancient oak trees that are over 1,000 years in age. Ultimately, we aim to understand how plant genomes are organised, expressed and how they evolve, leading to the immense phenotypic diversity that we see in the natural world.
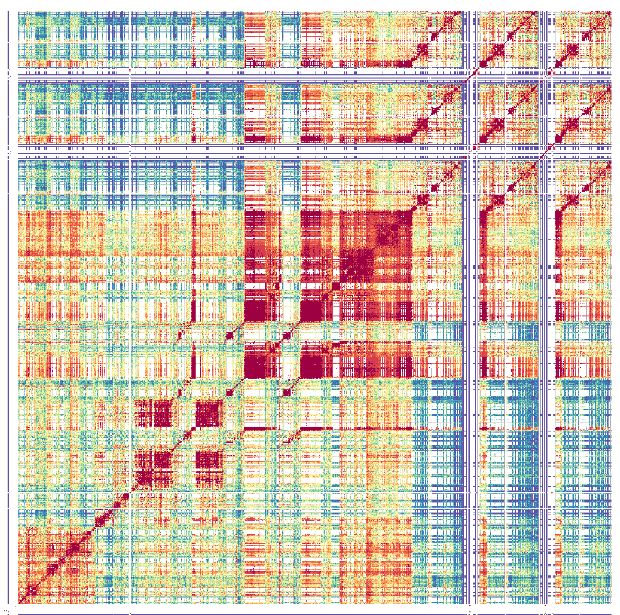
Caption: Patterns of sequence similarity within an Arabidopsis centromere. A heat map is shown where a single Arabidopsis centromere has been compared to itself. The red and orange colours indicate very high levels of self-similarity, whereas green and blue represent lower levels of similarity. The coloured patterns indicate complex patterns of sequence repetition within the centromere. Research in Prof. Henderson's group investigates how these patterns arise during evolution, and why the centromeres change so rapidly.