We use engineering approaches to understand biological complexity and advance plant biotechnology. Our research is focused on understanding how quantitative plant phenotypes emerge from network functions and exploring and utilising metabolic diversity. Our long-term goals are to develop the knowledge and technologies required to optimise crop performance through the rational engineering of regulatory networks and to provide routes for the sustainable use of natural products used in medicine, industry, and agriculture.
Understanding how phenotypes emerge from network functions
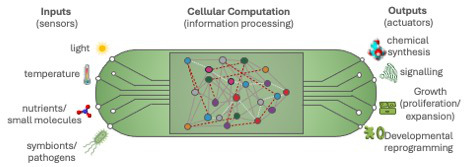
Quantitative traits are controlled by suites of genes working combinatorially within complex gene regulatory networks (GRNs). The complexity of GRNs has made them challenging to investigate using traditional genetic approaches. Similarly, predicting the effects of perturbations has been a significant barrier to applying genetic engineering to the improvement of quantitative traits.
Figure 1: The process of rewiring or reconstructing regulatory networks using synthetic biology approaches can aid our understanding of how phenotypes emerge from network functions.
The process of rewiring or reconstructing regulatory networks using synthetic biology approaches can aid the understanding of how phenotypes emerge from network functions and inform rational engineering. As GRNs are underpinned by interactions between transcription factors and regulatory elements, one focus of our work is to investigate these interactions and their impact on the intrinsic and emergent properties of cis-regulatory elements. Our long-term goals are to develop the knowledge and technologies required to optimise crop performance through the rational engineering of regulatory networks.
Exploring and utilising metabolic diversity
Plants produce a vast array of biologically active metabolites that help them adapt and survive in their ecological niches. Understanding the origin and evolution of plant metabolites is fundamental to explaining both the distribution of natural products among plant families and their biological roles. The richness of bioactive molecules found in plants also provides a wealth of potential pharmaceuticals, insecticides, flavours and fragrances and molecules of medicinal and industrial value.
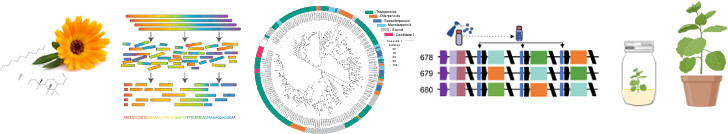
Figure 2: Identifying the genes involved the synthesis of natural products helps us to understand chemical diversity and enables the biomanufacturing of useful products for medicine and industry.
In the past decade, synthetic biology has accelerated methods for reconstructing the biosynthetic pathways of high-value natural products in so-called ‘chassis’ organisms. In our lab we integrate genomics, metabolomics, and bioactivity assays to identify molecules responsible for the bioactivity of plant extracts and to investigate the genetic basis of natural products. This enables us to understand mechanisms of metabolic diversification and explore methods for biomanufacturing. We also develop plants as photosynthetic biomanufacturing platforms, engineering synthetic circuits and tailoring plant genomes to optimise yield.
More information
To find out more about the research carried out by the Plant Molecular Engineering Research Group visit the Patron Lab website.