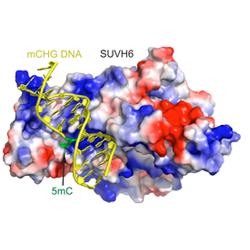
We are interested in how chromatin impacts gene expression. Plants are exquisitely attuned to changes in environmental conditions, such as temperature, light, drought and disease challenge. Chromatin helps establish long-term, sometimes multi-generational, cellular memories of events that modulate the transcriptional response to a renewed attack. This is akin to humans responding more effectively to a virus after natural or vaccine induced immunisation. In plants, such memories involve a re-sculpting of the chromatin landscape so that certain genes can be reactivated or repressed more rapidly with subsequent challenge. The major goal of our research is to discover how these mechanisms work, so that we can eventually control them to improve crop resistance and yield. This work aims to provide key insight and tools to address the global challenges of sustainable food production in the face of climate change and population growth.
Key Research Questions:
- How are chromatin and epigenetic marks perceived by the cell?
- What changes to chromatin occur during stress and memory formation?
- Can we develop CRISPR based approaches for epi-genome engineering?
To address these questions, we employ a range of molecular biology approaches, as well as cutting-edge (epi)genomic, proteomic and bioinformatic techniques to dissect chromatin memory and function.