Submitted by Anonymous on Wed, 17/05/2023 - 16:01
Centromeres are chromosome regions that play the same vital role during cell division across the tree of life, yet display enormous interspecies diversity. An international team of researchers has discovered that variation in centromere DNA sequences can be strikingly large even within a single species. These findings, now published in the journal Nature, shed light on the molecular mechanisms of rapid centromere evolution and their potential role in the formation of new species.
The centromere paradox
Each chromosome contains a special region called the centromere. During cell division, centromeres ensure that daughter cells receive the correct number of each chromosome. Despite their conserved role in cell division, centromeres of different species vary radically in size and structure. Biologists have termed this contrast between extreme diversity and conserved function the ‘centromere paradox. Team member Robin Burns notes, ‘While this paradox has been known for a long time, it has been difficult to explore due to the complexity of centromere DNA. We can now resolve this challenge using long-read sequencing technology’.
The centromere paradox inspired an international research team led by Ian Henderson (Department of Plant Sciences, University of Cambridge), Detlef Weigel (Max Planck Institute for Biology, Tübingen), and Alexandros Bousios (University of Sussex) to investigate plant centromere diversity. The researchers considered two alternative hypotheses: centromere diversity could be shared by all species and passed down from a common ancestor. Alternatively, it could be recent, emerging after species diverge.
Massive within species centromere diversity
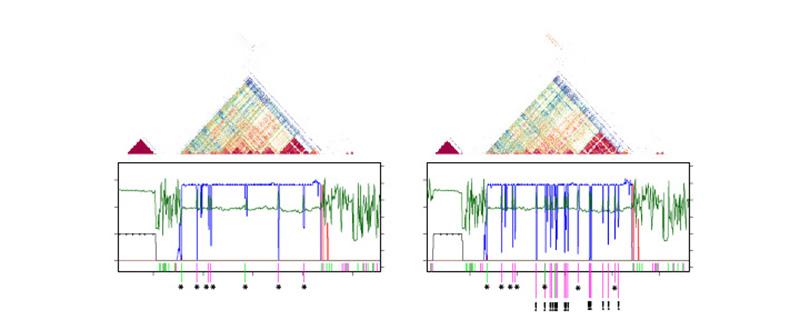
To find out, the team analyzed 66 individuals from Arabidopsis thaliana, a favourite model organism of plant scientists, as well as material from the sister species Arabidopsis lyrata. The team showed that even closely related individuals could have substantially different centromeres, despite all being built from the same family of short repeat units. ‘Our analysis proved both our alternative hypotheses true’, Weigel summarizes, ‘There is striking diversity within the species; but comparing the two species with each other, the differences look even more extreme’.
Invaders of the centromere
The dramatic centromere diversity observed raises questions about what could be responsible for rapid evolution. The team found that the Arabidopsis centromeres have been recently invaded by a mobile genetic element called ATHILA. This ‘selfish gene’ is a distant relative of retroviruses, such as HIV, yet lacks the ability to escape the cell. To counter ATHILA invasion, the cells actively overwrite the centromere repeats, thereby eliminating new ATHILA. Consequently, the small repeats that make up most of the centromere may be copied over-and-over again, causing them to diverge.
Team member Fernando Rabanal concludes, ‘It seems as if these repeated cycles of invasion and purging are the main drivers of rapid centromere evolution’. These mechanisms might be paralleled in other species, including humans. Bousios explains: ‘It is amazing to think that this cycle of hide and seek within centromeres may be repeated across species’, although he emphasizes, ‘The game remains the same, yet the actors are different’.
In the future, the team intends to deepen their understanding of rapid centromere change and how this may contribute to speciation. Henderson notes, ‘We have examined centromeres deeply within one species - in the future it will be important to study this kaleidoscopic diversity across the entire tree of life’. The questions sparked by the results are manifold, as team member Piotr Wlodzimierz points out: ‘We have yet to fully understand mechanisms of centromere repeat evolution. Recognizing their diversity is the first step in a journey that will illuminate the dark matter of a genome’.
Key contributors to the study include the groups of Polina Novikova (Max Planck Institute for Plant Breeding Research, Cologne, Germany), Martin Lysak (CEITEC, Brno, Czech Republic), Magnus Nordborg (Gregor Mendel Institute, Vienna, Austria), Fabrice Roux (CNRS, Toulouse, France), and Carlos Alonso-Blanco (Centro Nacional de Biotecnología, CSIC, Madrid, Spain).
The study was funded by the ERC, UKRI, HFSP, EMBO, the Max Planck Society, the Royal Society, the Czech Science Foundation, and the Ministerio de Ciencia e Innovación of Spain.
Text by Ian Henderson. Artwork by Ella Maru.
Read the paper
Wlodzimierz P*, Rabanal FA*, Burns R*, Naish M, Primetis E, Scott A, Mandáková T, Gorringe N, Tock AJ, Holland D, Fritschi K, Habring A, Lanz C, Patel C, Schlegel T, Collenberg M, Mielke M, Nordborg M, Roux F, Shirsekar G, Alonso-Blanco C, Lysak MA, Novikova P, Bousios A, Weigel D, Henderson IR. (2023) Cycles of satellite and transposon evolution in Arabidopsis centromeres. Nature. Vol, No. DOI: 10.1038/s41586-023-06062-z
*Equal contribution
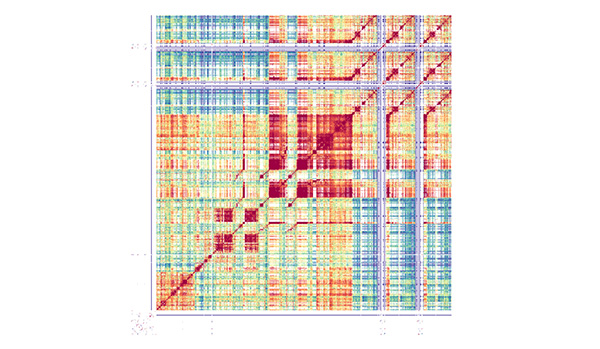
Above: A StainedGlass sequence identity heatmap of the centromere 5 satellite repeat array from the French MERE-A-13 accession. The red and orange shading indicates regions of high sequence similarity.