As part of our celebrations to mark 300 years since the appointment of the first Professor of Botany, some of our current academics have written short research stories to help give you an insight into current areas of interest and future research challenges.
If you are interested in finding out more, including how you might be able to support our academics in their future research endeavours, please get in touch with them directly.
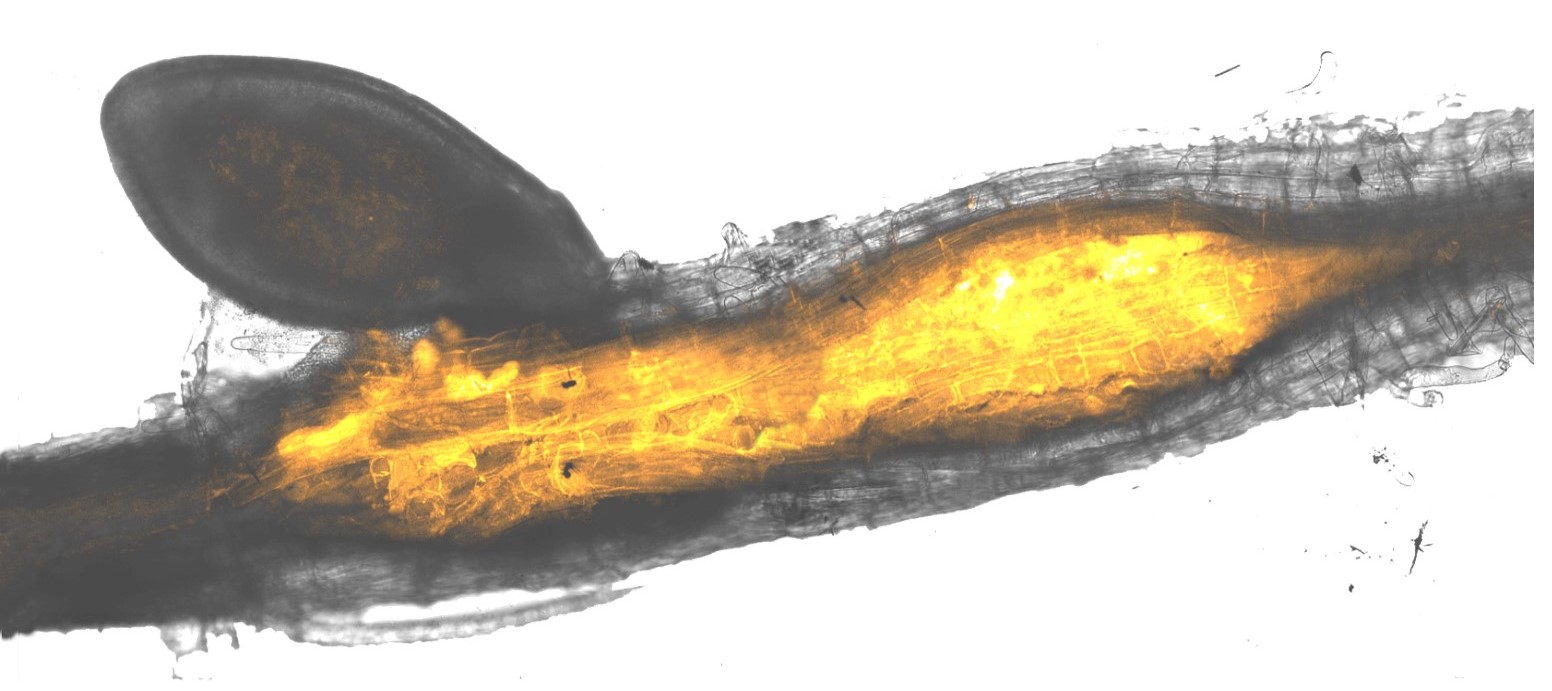
Know your enemy: discovery-driven sustainable solutions to plant-nematode interactions
Sebastian Eves-van den Akker, Head of Plant-Parasite/Pathogen Interactions Group
Problem we’re looking to solve
The problem we have chosen to work on is the molecular dialogue between two kingdoms of life: plants and their invading parasitic nematodes (microscopic worm-like creature). We are driven by fundamental discovery and, given that the outcome of this molecular dialogue results in plant disease that constrain human food security, we have a profound sense of responsibility to also deliver sustainable solutions to these parasites. To give an idea of scale, parasitic nematodes affect most crops, can cause up to 80% yield loss, and are estimated to account for $100 billion in annual loses to global agriculture.
Solution
It is widely accepted that plants which are inherently (i.e. genetically) resistant to a disease are an optimal solution in agriculture from practical, financial, and sustainability/equity-oriented points of view because no additional work/cost/inputs are required to prevent disease and thereby yield loss. Given the scarcity of such resistant plants, we focus on identifying, characterising, and ultimately understanding the plant genes that are responsible for resistance and susceptibility to disease.
Why Cambridge, why now, and history of working in this area
While Cambridge has a not insubstantial history of plant-nematode research (dating back some 60 years), the reason why our group is based here is because of critical mass and future direction. Our research group is based at the Crop Science Centre, a recent initiative between the Department of Plant Sciences and the National Institute of Agricultural Botany (NIAB) designed to deliver transformational change in agriculture, rooted in fundamental discovery. Principles of sustainability and equity in farming unite our collective aims. In addition to this impetus, the Crop Science Centre also houses a concentration of expertise on plant interactions with other organisms (ranging from pathogenic to beneficial) that is rare, if not unique. This allows us to share thinking and resources, drawing parallels across systems, to accelerate our collective research.
What is the research – what is it, how does it work, what resources are used and who is involved?
How you get to a resistant plant is important. There are, in essence, two ways to do this: the first, and most intuitive, is to add a gene to the plant that has some negative impact on the pathogen (often so called “resistance genes” or “R genes”); the second, and somewhat unintuitive, is to remove a gene from the plant that the pathogen was relying on manipulating in some way in order to cause disease (so called “susceptibility genes” or “S genes”).
The evolutionary biologist might favour the S gene because of a fundamental rule of nature: breaking things is easy - making things is hard. Intuitively, we all know this to be true without the proof. It is easier to break your phone, than it is to make yourself a new phone. The same is true with the interactions between proteins: mutations that change the shape of a protein are much more likely to break its function than they are to make a new function. This is important because many R genes added to plants work by recognising pathogen proteins in order to mount an immune response. Breaking this recognition is “easy”, and so pathogens readily evolve to avoid this type of resistance. On the other hand, resistance derived by removing an S gene from the plant, that the pathogen was manipulating and relying on, requires the pathogen to regain this ability in some way (i.e. making something new), and making things is “hard”. As a result, it is generally harder for pathogen evolution to overcome a resistant plant derived from the loss of an S gene, than it is for pathogen evolution to overcome a resistant plant derived from the addition of an R gene.
The biotechnologist and the politician might agree with the evolutionary biologist because of CRISPR/Cas genome editing (termed CRISPR). CRISPR is a revolutionary technology that allows precise edits in the genomes of almost any organism. CRISPR/Cas is extremely good at making targeted deletions in crop genomes, and its approval for commercial use in many geographies is streamlined compared to other biotechnological approaches to crop improvement (e.g. genetic modification). There is a clear opportunity to use CRISPR/Cas to develop disease resistant crops - based on fundamental understanding - provided the resistant plant is derived by making a targeted deletion (i.e. removing a gene). It is for this reason, that the biotechnologist, the politician, and the evolutionary biologist might favour the S gene.
Discovering more S genes is therefore paramount. We recently discovered a new way to find S genes – termed, “the hologenome theory of S gene discovery”. The term hologenome, coined for use in the hologenome theory of evolution, argues that we should consider the host plus all of its associated symbiotic microbes as a single entity, because they are inseparable in all practical senses. We used this definition to consider the host and the plant-parasitic nematode – a plant-pathogen of global agricultural importance – as a single unit. Conceptualising them in this way, we could ask, “what is the metabolic capacity of the plant-nematode hologenome?”. Identifying metabolic pathways that were in part contributed by the host genome and in part contributed by the parasite’s genome would highlight genes in the plant that the nematode relies on – and thereby new S genes for CRISPR-mediated crop improvement.
What are the milestones, timelines and what happens next?
Our current work in this area is progressing on two fronts: 1) Discovery of additional S genes based on hologenome theory; and 2) Translating these discoveries into crops using genome editing. Recent legislation (The Precision Breeding Bill) accelerates the approval process, so we anticipate rapid deployment of new material in field trials in the coming years.
If we solve the problem what can we expect
Most plants, and all crops, can be infected by one species of plant-parasitic nematode or another. These parasites have a profound impact on global agriculture – affecting both the richest and the poorest farming systems with equal vigor, but of course unequal outcomes. Addressing plant diseases is therefore a problem of not just sustainability, but also of equity.