Origins
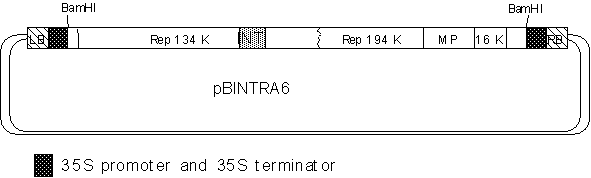
PTV00 is a tobravirus vector based on RNA 2 of TRV strain PPK20. This vector was derived from pCa7, a 35S driven full length infectious clone of PPK20 RNA 2 kindly provided by Prof J Bol (Hernandez et al., 1995) To construct pTV00 the modified pCa7 insert was transferred to the pGreen 0000 RLSK Agrobacterium binary vector (Hellens et al., 2000). PTV00 requires proteins encoded by RNA1 for replication and movement. PBINTRA6 is a Bin19 based construct containing 35S RNA 1 and an intron. The intron prevents expression of the replicase in E. coli and Agrobacterium and thereby promotes stability of the DNA. PBINTRA6 must be introduced to plants as well as pTV00 to obtain infection (Ratcliff et al., 2001).
Design
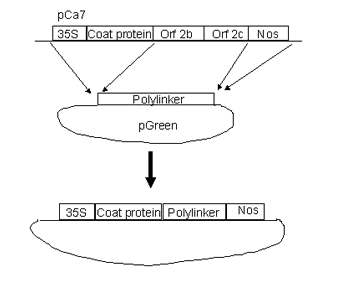
The DNA sequence of pTV00 is being determined and will be available from Genebank. Unique sites in polylinker are (5' to 3') SpeI, BamHI, SmaI, XmaI, HindIII, BspDI, ClaI, AccI, ApaI and KpnI.
The sequence of pBINTRA6 is in GeneBank (accession number #AF314165)
Purpose
Tobravirus derived vectors have several potential advantages over other currently available virus vectors.
- The presence of a large polylinker of unique sites, and the small overall size of the vector should facilitate cloning of inserts.
- Virus infections that lack Orfs 2b and 2c are symptomless in N. benthamiana and very mild in Arabidopsis; the effects of protein expression or VIGS are therefore clearer in the absence of virus symptoms.
- Tobraviruses are unusual in infecting meristems, floral organs and even pollen. pTV should therefore be able to express or silence genes in these areas.
- TRV does not block the systemic signal for silencing. Even uninfected cells may therefore become silenced.
Use
The vectors are provided as DNA samples. To propagate in E. coli use 50µg/ml kanamycin for both constructs. We recommend E. coli strain DH5a, especially for pBINTRA6, since it tends to be unstable in E. coli. For propagation in A. tumefaciens use C58c1 for pBINTRA6 and GV3101 with pSa-rep (Hellens et al., 2000) for pTV00 and derivatives and select with 50µg/ml Kan and 5 µg/ml Tet and 50µg/ml Rifampicin
N. benthamiana
Separate agro cultures (10 ml) containing pBINTRA6 and the TRV RNA2 constructs are grown up overnight, spun down at 3k rpm for 20 mins, and resuspended in the same volume of 10µM MgCl2, with 100µM acetosyringone (in DMSO) and 1µM MES pH 5.6. These cultures can be mixed at any point after re-suspension (ie in the lab).
We infiltrate young leaves. Temperature should be below 24C, since temperatures above it may inhibit TDNA transfer.
Originally we infiltrated Agro containing RNA 1 and RNA 2 in a 1:1 ratio. However, surprisingly we can obtain the same VIGS if LESS RNA 2 (but not less RNA1) is used.
Arabidopsis
The TRV vector is not completely efficient in Arabidopsis. Typically we observe VIGS in 10-80% of the inoculated plants. VIGS is normally transient between 10 dpi and 3-4 weeks post inoculation although it does persist sometimes.
For infection of Arabidopsis we first inoculate N. benthamiana by infiltratio and extract sap from infiltrated leave at 3 days post infiltration (dpi). The sap is then inoculated directly to Arabidopsis. Work to improve this system is ongoing. In some experiments we purify virions using the following procedure.
- take infiltrated leaves.
- Grind/crush/smash them thoroughly in 4 ml 100mM Phosphate buffer pH 7.2 (Manniatis). -Freeze -Thaw (breaks open more cells)
- Spin 9000 g for about 1 hour. Keep supernatant.
- Filter through fine filter (we use Mira cloth)
- add 10%wv PEG 6000 and 0.34 M NaCl. Dissolve, then keep on ice at 4c over night (precipitates virus).
- Spin at 9000g for 1 hour. Keep pellet.
- Resuspend pellet in about 0.5 ml phosphate buffer (we vortex for about an hour in our cold room).
- Spin down any lumps; they are just rubbish.
- lightly dust your plants with Carborundum grinding powder.
- rub about 5ul of virus prep onto 3 leaves per plant. Small (2-3 week) arabs, not old cabbages.
Controls
Favourite control of the day is pTV09, which contains the sulphur gene. This gene sequence is very conserved and so pTV09 works on benths and Arabs. Sulphur is a magnesium chelatase required for chlorophyll production, and the plants turn yellow without it (Kjemtrup et al., 1998).
Nomenclature
The empty vector, containing no subgenomic promoter and no insert, is called pTV00. In this system, the first figure represents the presence and nature of a subgenomic promoter, while the second represents the presence and nature of the insert. Each insert should therefore have a unique number, as listed below.
Primers
To facilitate PCR colony-screening, forward and reverse primers have been designed side of the polylinker. The empty pTV00 vector gives a PCR product of 350bp. Primers are:
FR69 pTV00 forward primer: 5' gctgctagttcatctgcac
FR70 reverse primer for pTV00 5'gcacggatctacttaaagaac
These primers are optimum at 53°c. M13 forward is also present in reverse orientation, downstream of the polylinker (ie as reverse). This is useful for PCR screening colonies, but is repeated further downstream, so is useless for sequencing.
References
Hellens, R.P., Edwards, E.A., Leyland, N.R., Bean, S.and Mullineaux, P.M. (2000). pGreen: a versatile and flexible binary Ti Vector for Agrobacteruim-mediated plant transformation. Plant Mol. Biol. 42, 819-832.
Hernandez, C., Mathis, A., Brown, D.J.F.and Bol, J.F. (1995). Sequence of RNA 2 of a nematode transmissible isolate of tobacco rattle virus. J. Gen. Virol. 76, 2847-2851.
Kjemtrup, S., Sampson, K.S., Peele, C.G., Nguyen, L.V., Conkling, M.A., Thompson, W.F.and Robertson, D. (1998). Gene silencing from plant DNA carried by a geminivirus. Plant J. 14, 91-100.
Ratcliff, F., Martin-Hernandez, A.M.and Baulcombe, D.C. (2001). Tobacco rattle virus as a vector for analysis of gene function by silencing. Plant J. 25, 237-245.
