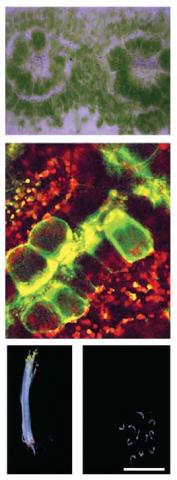
We are interested in understanding photosynthesis and developing strategies that may allow it to be improved in the future. Our major focus relates to how the efficient C4 pathway has evolved from the ancestral C3 state. In almost all C4 plants, leaves are modified such that changes to their development, cell biology and biochemistry allow photosynthesis to be partitioned between two cell-types. Despite this complexity, C4 photosynthesis has arisen in over sixty separate lineages of angiosperms, and compared with the C3 pathway, photosynthetic efficiency is increased by around 50%.
Our work has provided a number of insights that help explain how C4 photosynthesis operates and evolved. For example, we demonstrated that C3 plants possess cells in stems and petioles that carry out a rudimentary C4 cycle; we isolated the first cis-elements that control the expression of multiple photosynthesis genes in either mesophyll or bundle sheath cells of C4 leaves, and showed that these regulators are found in the ancestral C3 leaf; we provided the first estimates of the extent to which homologous transcription factors are recruited into independent C4 lineages; and we provided the first direct evidence that duons (transcription factor binding sites that reside in exons impact on gene expression) – again these duons are present in the ancestral C3 state. We continue to investigate the regulation of photosynthesis gene expression in C3 and C4 leaves to better understand the molecular parts that evolution has excapted. Projects include:
Our work focusses on two complementary systems that in both cases use C3 leaves to provide a foundation for our understanding of the derived C4 system.
First, using the C3 model Arabidopsis thaliana and its closest C4 relative Gynandropsis gynandra, we aim to understand how the regulation of photosynthesis gene expression has altered in specific cell types of C4 compared with C3 dicotyledenous leaves. Here we make use of molecular biology, genome-wide analysis of gene expression and transcription factor binding but also natural variation found within a species.
Second, using rice, we wish to better understand how photosynthesis is regulated in specific leaf cells of this important monocotyledonous crop. In particular, we are interested in why, in contrast to the mesophyll cells of rice, the bundle sheath cells do not fill up with chloroplasts. Here, we make use of molecular biology, genome-wide analysis of gene expression and transcription factor binding in particular cell-types, and synthetic biology approaches to probe and engineer the rice leaf.
More information
To find out more about the research carried out by the Molecular Physiology Research Group visit the Hibberd Lab website.
Images (from top): Kranz anatomy of a C4 leaf; Bundle sheath cells marked with reporter; Knock out mutants of C3 Arabidopsis lacking proteins recruited into C4 photosynthesis are compromised in early seedling growth.