Head of Group: Sir David Baulcombe
Edward Penley Abraham Royal Society Research Professor and Regius Professor of Botany Emeritus
Unfortunately the group is no longer accepting applications for studentships or scientific visitors. Postdoctoral vacancies and other vacancies will be advertised here as they arise.
Our research:
Understanding the growth, development, physiology and evolution of plants depends on knowing how genes are switched on and off. Progress in crop improvement relies similarly on having unravelled networks of genetic regulation. To shed light on these processes we investigate interspecific hybrids and disease resistance in tomato.
The unifying themes in these projects derive from previous discoveries in small regulatory RNA, epigenetics and the mechanisms of disease resistance in plants. An overview of these past discoveries is described in David Baulcombe's brief memoir entitled ""
Current projects include:
Disease and disease resistance
Our earlier research into disease and disease resistance had two separate strands based on RNA and protein based mechanisms. In our RNA work we discovered small interfering (si)RNAs with many diverse roles including anti-viral defense. The protein work focused on innate immunity proteins with nucleotide binding sites and leucine rich repeats (NLRs). We demonstrated how the domains of these proteins may interact intra-molecularly (Moffett et al, 2002), how these proteins interact inter-molecularly with each other (Peart et al, 2005) and how they can be evolved artificially to have novel disease resistance specificity (Farnham & Baulcombe, 2006; Harris et al, 2013).
A recent finding brings these two strands together because NLR mRNAs are regulated by a cascade involving siRNAs (Shivaprasad et al, 2012a). If this cascade is blocked we can enhance quantitative disease resistance to Phytophthora infestans (an oomycete) and Pseudomonas syringae (a bacterium) (Canto-Pastor et al, 2019).
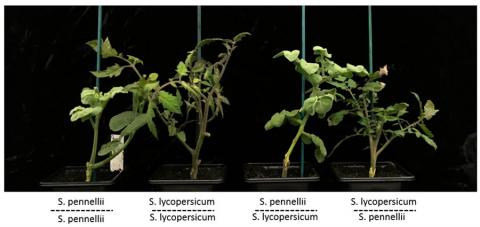
Interspecific hybrids
Our earlier discovery of a link between siRNA and epigenetics (Herr et al, 2005) prompted the hypothesis to explain why interspecific hybrids are often much better or much worse than either of the parents. We proposed that siRNAs may mediate interactions between genomes in these hybrids so that there are new patterns of gene expression that are unlike either of the parents.
 To test this hypothesis we are following hybrids of cultivated and wild tomato species through to the F4 generation and have evidence that is supportive of the hypothesis (Shivaprasad et al, 2012b).
To test this hypothesis we are following hybrids of cultivated and wild tomato species through to the F4 generation and have evidence that is supportive of the hypothesis (Shivaprasad et al, 2012b).
The inheritance pattern of some epigenetic changes in these hybrid plants did not always follow Mendelian ratios and, in some instances, there was evidence of a "gene drive" in which an epigenetic silenced state is transferred from one allele to another. This process resembles paramutation that has been characterised previously in maize (Gouil & Baulcombe, 2018, 2016).
Our interest in epigenetics takes us into the world of transposons and, from our recent analysis (Wang and Baulcombe 2020), it seems that recent and ancient transposons are silenced through a process that requires siRNA but those in the 'middle aged' category are silenced in an RNA-independent manner. Our ageing hypothesis to explain these findings proposes that RNA-based silencing is the first line of defense against transposons. The RNA-independent mechanism is more secure and is effective through the period when transposons acquire mutations and eventually lose the ability to transpose and mutate the genome. In the late phase the transcriptional suppression of non-functional transposons can be relaxed without jeopardising genome integrity. Any siRNAs derived from the transcripts would have a genome protection role because they would silence new transposons with sequence similarity to these late phase elements in the genome.
We can also explore the interaction between genomes in chimaeric grafts rather than by sexual crossing because siRNA can move across a graft union and direct epigenetic change in the recipient tissue. Our initial characterisation of this mobile siRNA was in Arabidopsis (Lewsey et al, 2016; Molnar et al, 2010; Melnyk et al, 2011) and we are now testing the system in tomato.
Research methodology
 We rely heavily on next generation sequencing and, consequently, on bioinformatics (include link to bioinformatics page from current website). Gene editing has allowed us to introduce mutations into genes of interest in tomato and we also use virus-induced gene silencing. We are exploring the use of virus and other vectors to introduce the guide RNA in gene editing in N. benthamiana.
We rely heavily on next generation sequencing and, consequently, on bioinformatics (include link to bioinformatics page from current website). Gene editing has allowed us to introduce mutations into genes of interest in tomato and we also use virus-induced gene silencing. We are exploring the use of virus and other vectors to introduce the guide RNA in gene editing in N. benthamiana.
Impact
Our research can be used to explain the concepts of epigenetics, regulation and disease resistance in plants. At a practical level we aim to develop improved strategies in crop plant breeding and broad spectrum disease resistance in crops. We are also exploring the potential of epigenetically modified crops.
David Baulcombe led the fundraising for a crop science initiative with NIAB and other colleagues in the University. This Crop Science Centre will facilitate collaboration in translation of research by University researchers and also by the Russell R. Geiger Professor of Crop Science.
References
- Canto-Pastor A, Santos B, Valli A, Summers W, Schornack S & Baulcombe D (2019) Enhanced resistance to bacterial and oomycete pathogens by short tandem target mimic RNAs in tomato. PNAS: 396564 Available at: www.pnas.org/cgi/doi/10.1073/pnas.1814380116
- Farnham G & Baulcombe D (2006) Artificial evolution extends the spectrum of viruses that are targeted by a disease-resistance gene from potato. Proc. Natl. Acad. Sci. U.S.A. 103: 18828–18833
- Gouil Q & Baulcombe DC (2016) SLTAB2 is the paramutated SULFUREA locus in tomato. J Exp Bot 67: 2655–2664
- Gouil Q & Baulcombe DC (2018) Paramutation-like features of natural epialleles in tomato. BMC Genomics 19: doi.org/10.1186/s12864-018-4590-4
- Harris CJ, Slootweg EJ, Goverse A & Baulcombe DC (2013) Stepwise artificial evolution of a plant disease resistance gene. Proc. Natl. Acad. Sci. U. S. A. 110: 21189–94
- Herr AJ, Jensen MB, Dalmay T & Baulcombe DC (2005) RNA polymerase IV directs silencing of endogenous DNA. Science. 308: 118–120
- Lewsey MG, Hardcastle TJ, Melnyk CW, Molnar A, Valli A, Urich MA, Nery JR, Baulcombe DC & Ecker JR (2016) Mobile small RNAs regulate genome-wide DNA methylation. Proc. Natl. Acad. Sci. U. S. A. 113: E801-810
- Melnyk CW, Molnar A, Bassett A & Baulcombe DC (2011) Mobile 24 nt small RNAs direct transcriptional gene silencing in the root meristems of Arabidopsis thaliana. Curr. Biol. 21: 1678–1683
- Moffett P, Farnham G, Peart JR & Baulcombe DC (2002) Interaction between domains of a plant NBS-LRR protein in disease resistance-related cell death. EMBO J. 21: 4511–4519
- Molnar A, Melnyk CW, Bassett A, Hardcastle TJ, Dunn R & Baulcombe DC (2010) Small Silencing RNAs in Plants Are Mobile and Direct Epigenetic Modification in Recipient Cells. Science. 328: 872–875
- Peart J, Mestre P, Lu R, Malcuit I & Baulcombe D (2005) NRG1, a CC-NB-LLR protein mediates resistance against tobacco mosaic virus together with N, a TIR-NB-LRR protein. Curr. Biol. 15: 968–973
- Shivaprasad P V., Chen H-M, Patel K, Bond DM, Santos BACM & Baulcombe DC (2012a) A microRNA superfamily regulates nucleotide binding site-leucine-rich repeats and other mRNAs. Plant Cell 24: 859–74
- Shivaprasad P V, Dunn RM, Santos BACM, Bassett A & Baulcombe DC (2012b) Extraordinary transgressive phenotypes of hybrid tomato are influenced by epigenetics and small silencing RNAs. EMBO J. 31: 257–66
- Wang, Z. & Baulcombe, D. C. Transposon age and non-CG methylation. Nat. Commun. 1–9 (2020). doi:10.1038/s41467-020-14995-6