Supervisor
Dr Jake Harris
Brief Summary
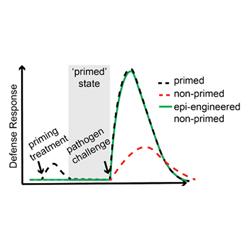
Plants can become more resilient to pathogen challenge after priming treatments, which is associated with changes to chromatin. This project uses epigenome engineering to re-sculpt chromatin to improve performance under pathogen pressure.
Importance of Research
Epigenome engineering aims to harness the plants natural defense and memory systems to improve resilience.
Project Summary
Plants defend themselves against pathogens by activating a suite of antimicrobial defense genes. Interestingly, plants exposed to certain pathogens can become more resistant to subsequent pathogen challenge, indicating that a type of immunity has been formed. In this ‘primed’ state, key antimicrobial defense genes are turned on more rapidly as compared to non-primed ‘naïve’ plants, which contributes to their heightened resistance. However, it is currently unknown how this elevated transcriptional response is achieved. Recent research suggests that epigenetic changes in chromatin may help form the basis for this immunological memory. For instance, particular chromatin marks can be laid down that help recruit transcriptional machinery. However, precisely which chromatin features drive priming, and how they achieve their function, remains mysterious. This project aims first to understand the pathogen primed state.
What will the successful application do?
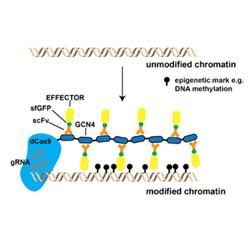
The project will consist of three stages. In the first stage, genome-wide chromatin profiling by ChIP-seq/CUT&Tag and ATAC-seq will be used to identify the chromatin features of primed vs non-primed plants. In the second stage, epigenome engineering tools based on the recently described CRISPR system, will be used to deposit chromatin marks at precise genomic locations. The final stage will investigate whether these epigenome-engineered plants are more resistant to pathogens.
Training Provided
The student will gain expertise in a range standard molecular biology techniques (cloning, genotyping, qRT-PCR, western blots). Additionally, the student will help to develop cutting-edge CRISPR based epigenome engineering tools, and will learn genomics (wet lab and computational approaches), and plant pathology techniques.
References
- Conrath U, Beckers GJ, Langenbach CJ, Jaskiewicz MR. (2015). Priming for enhanced defense. Annu Rev Phytopathol. 53:97-119. doi: 10.1146/annurev-phyto-080614-120132. Epub 2015 Jun 11. PMID: 26070330.
- Bäurle I, Trindade I. (2020). Chromatin regulation of somatic abiotic stress memory. J Exp Bot. Aug 17;71(17):5269-5279. doi: 10.1093/jxb/eraa098. PMID: 32076719.
- Papikian A, Liu W, Gallego-Bartolomé J, Jacobsen SE. (2019). Site-specific manipulation of Arabidopsis loci using CRISPR-Cas9 SunTag systems. Nat Commun. Feb 13;10(1):729. doi: 10.1038/s41467-019-08736-7. PMID: 30760722; PMCID: PMC6374409
- Harris CJ, Scheibe M, Wongpalee SP, Liu W, Cornett EM, Vaughan RM, Li X, Chen W, Xue Y, Zhong Z, Yen L, Barshop WD, Rayatpisheh S, Gallego-Bartolome J, Groth M, Wang Z, Wohlschlegel JA, Du J, Rothbart SB, Butter F, Jacobsen SE. (2018). A DNA methylation reader complex that enhances gene transcription. Science. Dec 7;362(6419):1182-1186. doi: 10.1126/science.aar7854. PMID: 30523112; PMCID: PMC6353633.